- What is the CGI?
- How does the CGI work?
- How are cancer genes identified?
- Which type of alterations does the CGI interpret?
- How are the driver events identified?
- What is BoostDM?
- How does BoostDM perform in the identification of driver mutations in cancer genes?
- What is OncodriveMUT?
- What is the performance of OncodriveMUT in the identification of driver mutations?
- How are mutations annotated with prior knowledge?
- How are actionable events identified?
- What is the Cancer Biomarkers database?
- What other resources are used to identify potentially actionable events?
- Can CGI be used to interpret more than one tumor sample at a time?
- Why is it necessary to select a cancer type when interpreting a sample(s)?
- Why is there a cancer taxonomy?
- How are the mutations mapped?
- How is the CGI output provided?
- Which is the CGI license?
- How to cite CGI?
- What is the input to run the CGI analysis?
- Which mutations formats are accepted?
- Which human genome assemblies are accepted?
- How long does it take to have my CGI analysis finished?
- Are there any platform requirements for the mutation data?
- What happens if a mutation can not be mapped to the reference genome?
- Are any mutations not analyzed/included in the report?
- Do I need to login to run a CGI analysis?
- Can I download CGI results as a file?
- Are my CGI results private?
- Can I share my CGI results?
- Can I provide feedback on CGI?
- Is there a REST API?
- How to access old versions of the CGI
What is the CGI?
Cancer Genome Interpreter (CGI) is designed to support the identification of tumor alterations that drive the disease and/or which may be therapeutically actionable. CGI relies on computational methods --In silico saturation mutagenesis of cancer genes (BoostDM and OncodriveMut)-- as well as on knowledge collected across the public domain to annotate the alterations in a tumor according to several levels of evidence.
How does the CGI work?
With a list of genomic alterations in a tumor of a given cancer type as input, the CGI automatically recognizes the format, remaps the variants as needed and standardizes the annotation for downstream compatibility. Next, it identifies the likely driver alterations in cancer driver genes in the tumor type in question using computational methods (BoostDM and OncodriveMut). Moreover, mutations observed in the tumor that are known to be tumorigenic (known oncogenic mutations) are annotated. Alterations that constitute biomarkers of response to anti-cancer drugs are identified according to several databases (CIViC, OncoKB and the Cancer Biomarkers database).

Schema summarizing the CGI framework
How are cancer genes identified?
The list of cancer genes across 66 tumor types were obtained from the systematic analysis of more than 28,000 tumor samples using the IntOGen pipeline.
Which type of alterations does the CGI interpret?
CGI analyses mutations (single nucleotide changes and small insertions/deletions), copy number alterations (gene amplifications and deletions) and translocations.
How are the driver events identified?
Driver single nucleotide variants are identified by boostDM, a machine learning-based method employed in the in silico saturation mutagenesis of cancer genes, or --if no model is available for a cancer gene in the tumor type of interest-- by OncodriveMut. Driver indels are identified by OncodriveMut. The identification of potential driver structural variants in the tumor, including Copy Number Alterations (CNA) and translocations, relies on prior knowledge accumulated over the years by cancer researchers. Specifically, we assessed the coherent change in expression of known CNA drivers from the Cancer Gene Census across 34 tumor types. Amplification drivers with a significant increase in expression (and deletion drivers with a significant decrease in expression) are annotated as ‘predicted’ drivers in the tumor type in question. Oncogenic translocation events, manually curated from diverse sources are also annotated.
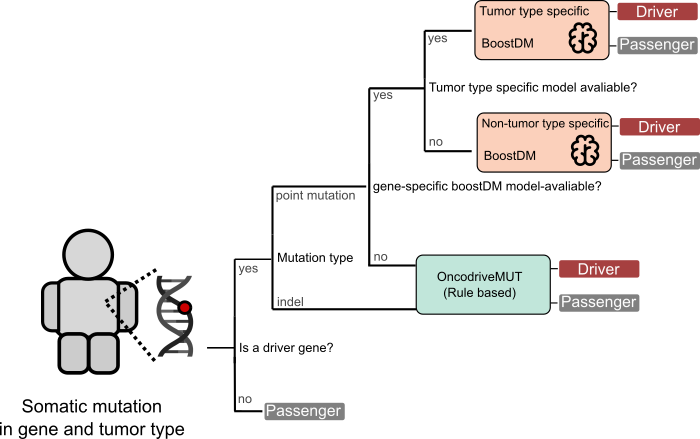
Identification of driver events
What is BoostDM?
BoostDM consists of an array of machine learning-based models trained for the identification of driver mutations affecting cancer genes. Briefly, a specific model is trained for a cancer gene in a tissue (given that a representative set of mutations has been observed across cohorts of tumors of the cancer type in question) which reflects the mechanisms of tumorigenesis affecting the gene in the tissue. Mutations observed across cohorts of tumors collected in IntOGen and a set of mutational features of cancer genes detected through the calculation of signals of positive selection are used to train the models. These models are then used to carry out an in silico saturation mutagenesis --i.e., classifying all possible single nucleotide variants as drivers or passengers-- of cancer genes. The classification across cancer genes and cancer types is used as the basis for the interpretation of single nucleotide variants in CGI. If a model is not available for a cancer gene in the tumor type of interest of the user, a model trained pooling mutations from closely related tumor types, or a model in a different tumor type are used.
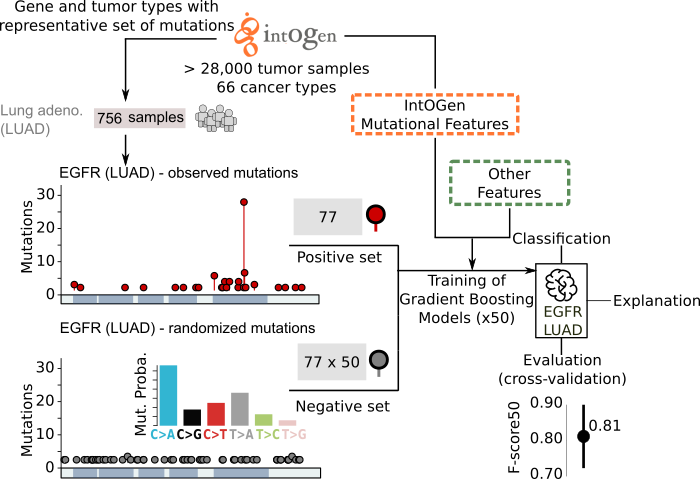
BoostDM
Importantly, an easy-to-read interpretation is provided for the classification of each mutation as driver or passenger. The radial plot illustrating this interpretation presents the contribution of each feature to the classification of the mutation, providing a rationale for the tumorigenic mechanism underpinning the driver mutation.

BoostDM
How does BoostDM perform in the identification of driver mutations in cancer genes?
Only high-quality boostDM models are used. Employing the mutations observed in a cancer gene across tumors as a proxy set of the driver mutations affecting the gene in the tissue in question, we demonstrated that boostDM models outperform several experiments of saturation mutagenesis and bioinformatic approaches.
How does BoostDM perform in the identification of driver mutations in cancer genes?
BoostDM models show a high F-score50 (above 90% in most cases) in the classification of two sets of experimentally validated oncogenic mutations observed across tumors.
How does BoostDM perform in the identification of driver mutations in cancer genes?
Finally, an evaluation of boostDM models in the real-life task of identifying unseen mutations across individual tumor samples (similar to their use in CGI) yielded performance comparable or better than those observed in the cross-validation.
These and other validation results are available at (link to the paper after publication).
What is OncodriveMUT?
OncodriveMut is a bioinformatics method to identify the most likely driver mutations of a tumor. Specifically, it classifies mutations observed in cancer genes (list) into drivers or passengers. Its main innovation with respect to other existing tools with a similar purpose is the incorporation of features characterizing the genes (or regions within genes) where the mutations occur, derived from the analysis of cohorts of tumors (6,792 samples across 28 cancer types) and samples from healthy donors (60,706 unrelated individuals). This knowledge is combined with features that describe the impact of the mutation on the function of the protein it affects via a set of heuristic rules to predict the effect of the mutations of uncertain significance.
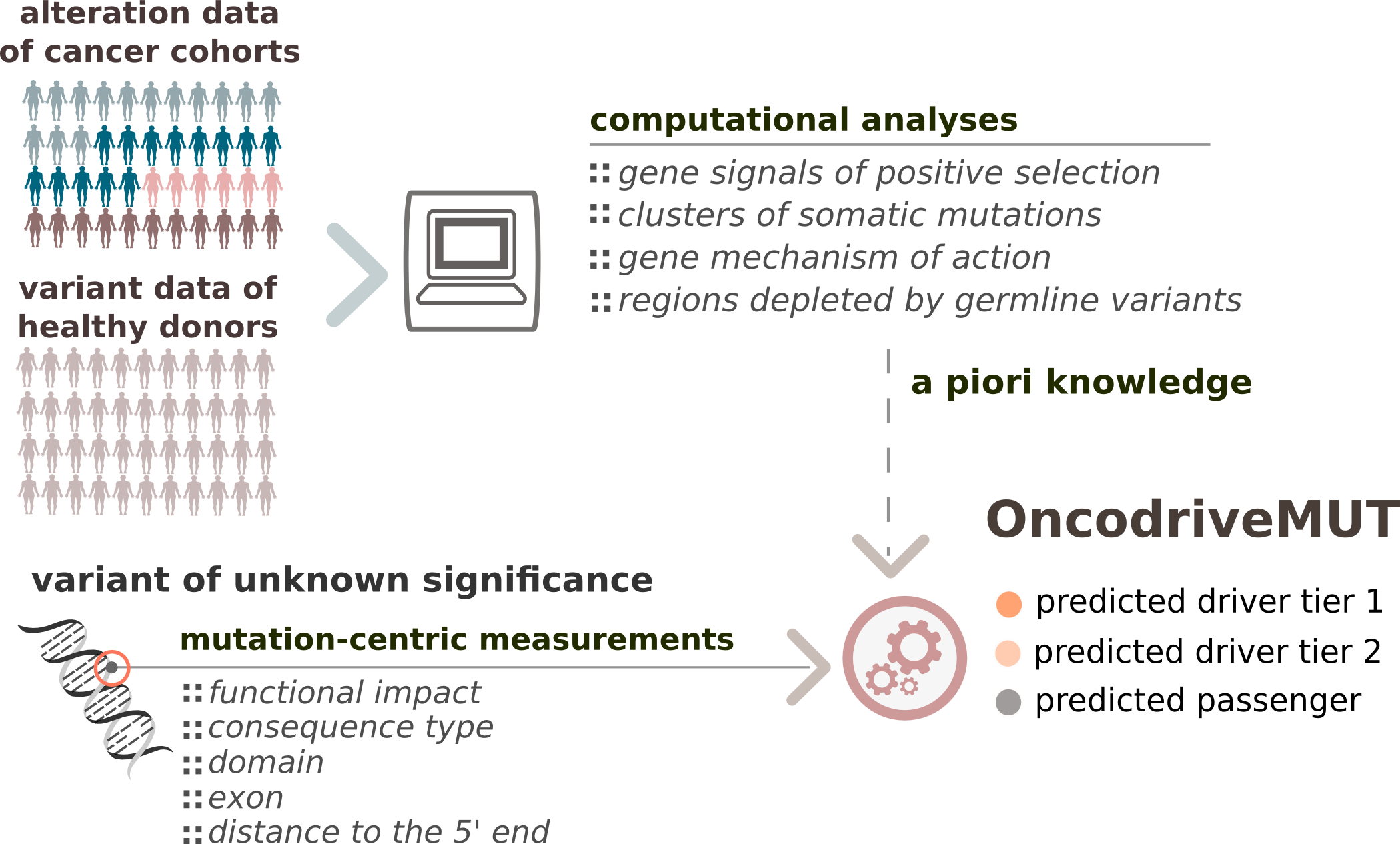
Schema summarizing the OncodriveMUT method framework
What is the performance of OncodriveMUT in the identification of driver mutations?
To benchmark the performance of OncodriveMUT, we compiled a set of known driver somatic mutations (n=1,077) and cancer-predisposing germline variants (n=2,819) (positive set) and another set of validated non-pathogenic somatic mutations (n=241) and common (major allele frequency > 1%) polymorphisms (n=1,006) affecting cancer genes (negative set). Noteworthy, only protein-affecting mutations in cancer genes have been included, a ground in which the distinction between driver and passenger mutations is more challenging than across all human genes. We found that OncodriveMUT distinguishes between the positive and negative sets of variants in cancer genes with an accuracy of 0.91 and Matthews correlation coefficient of 0.78.
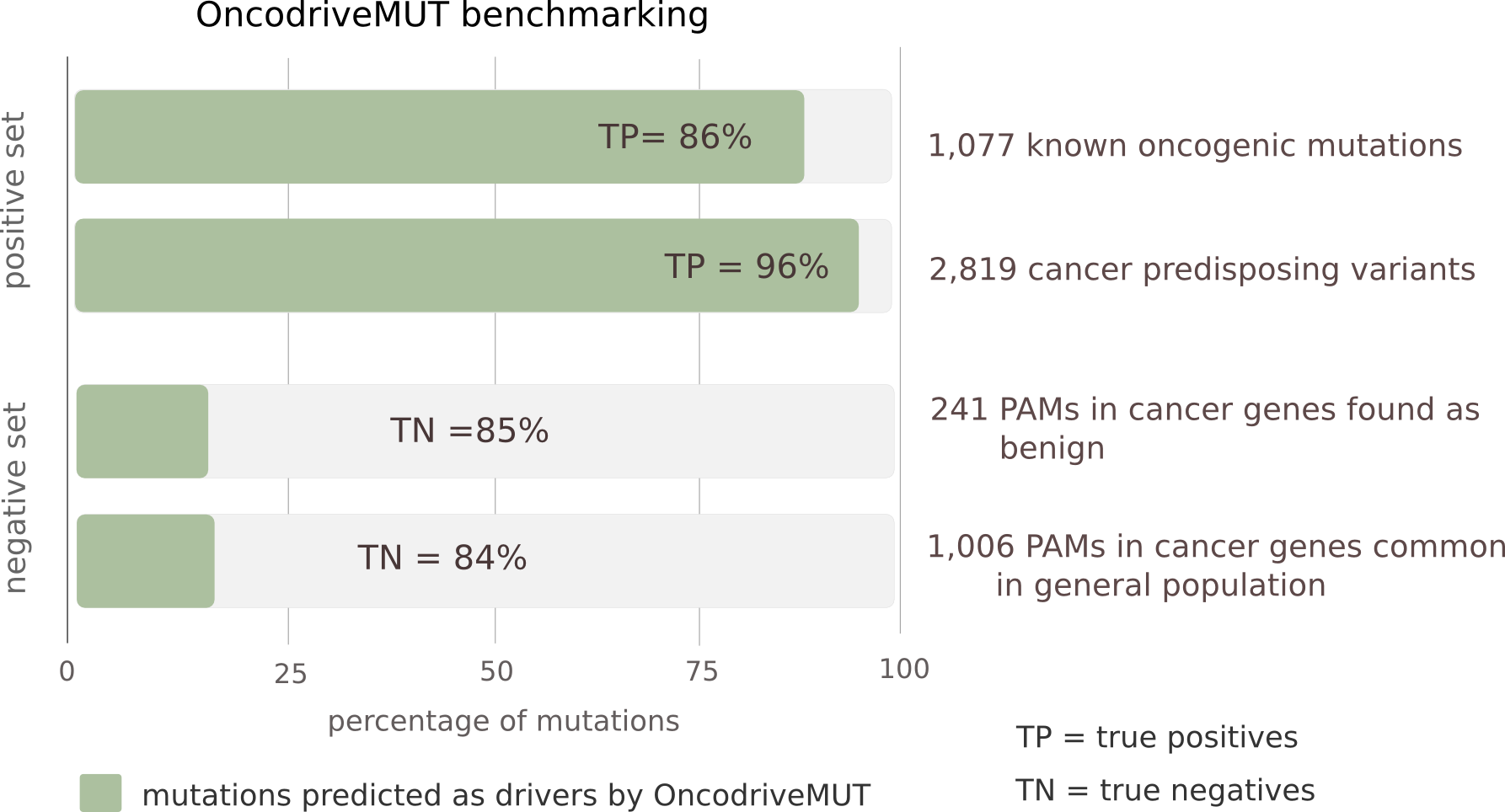
Performance of OncodriveMUT to distinguish bona fide driver and passenger protein-affecting mutations (PAM) among cancer genes
How are mutations annotated with prior knowledge?
Three repositories of mutations with relevance in tumorigenesis, collected across the scientific literature, are used to annotate mutations observed in the tumor. These are OncoKB, ClinVar, and our in-house set of validated oncogenic mutations. From ClinVar, somatic variants with clinical significance containing the terms ‘pathogenic’, ‘likely pathogenic’, ‘benign’ or ‘likely benign’ are retrieved.
How are actionable events identified?
To assess the relevance of the alterations as biomarkers of drug response, the CGI relies on an in-house database (Cancer Biomarkers-db) and two other publically available resources (CIViC and OncoKB) obtained via the Variants Interpretation in Cancer Consortium (VICC) of genomic events that influence the response of the tumor to a drug (sensitivity, resistance or severe toxicity). Thwith varying levels of clinical relevance according to state-of-the-art knowledge, ranging between standard-of-care guidelines and the evidences derived from preclinical assays.
In a previous version of the CGI, we also matched driver mutations in tumors with compounds that bind altered genes (Cancer Bioactivities-db). This is no longer supported.
What is the Cancer Biomarkers database?
The Cancer Biomarkers db integrates manually collected genomic biomarkers of drug sensitivity, resistance and severe toxicity. These biomarkers are classified by the cancer type in which they have been described according to different levels of clinical evidence supporting the association. The database is available for access and feedback by the community at www.cancergenomeinterpreter.org/biomarkers. The aggregation, curation and interpretation of the biomarkers follow the standard operating procedures developed under the umbrella of the H2020 MedBioinformatics project, thus ensuring the mid-term maintenance of these resources. The feedback from the community is also facilitated through the CGI web interface. Nevertheless, access to this type of data is both crucial for the advance of cancer precision medicine and highly complex to be comprehensively covered and updated by a single institution.
The Variant Interpretation for Cancer Consortium (VICC), under the Global Alliance for Genomics & Health framework (Global Alliance for Genomics and Health et al. 2016), has developed a Meta-Knowledgebase of biomarkers of tumors response to drugs. Given its completeness and the fact that it combines several dedicated resources (including the Cancer Biomarkers database), in the current version of Cancer Genome Interpreter (and in future releases) we rely on this Meta-Knowledgebase for the identification of biomarkers of response to drugs.
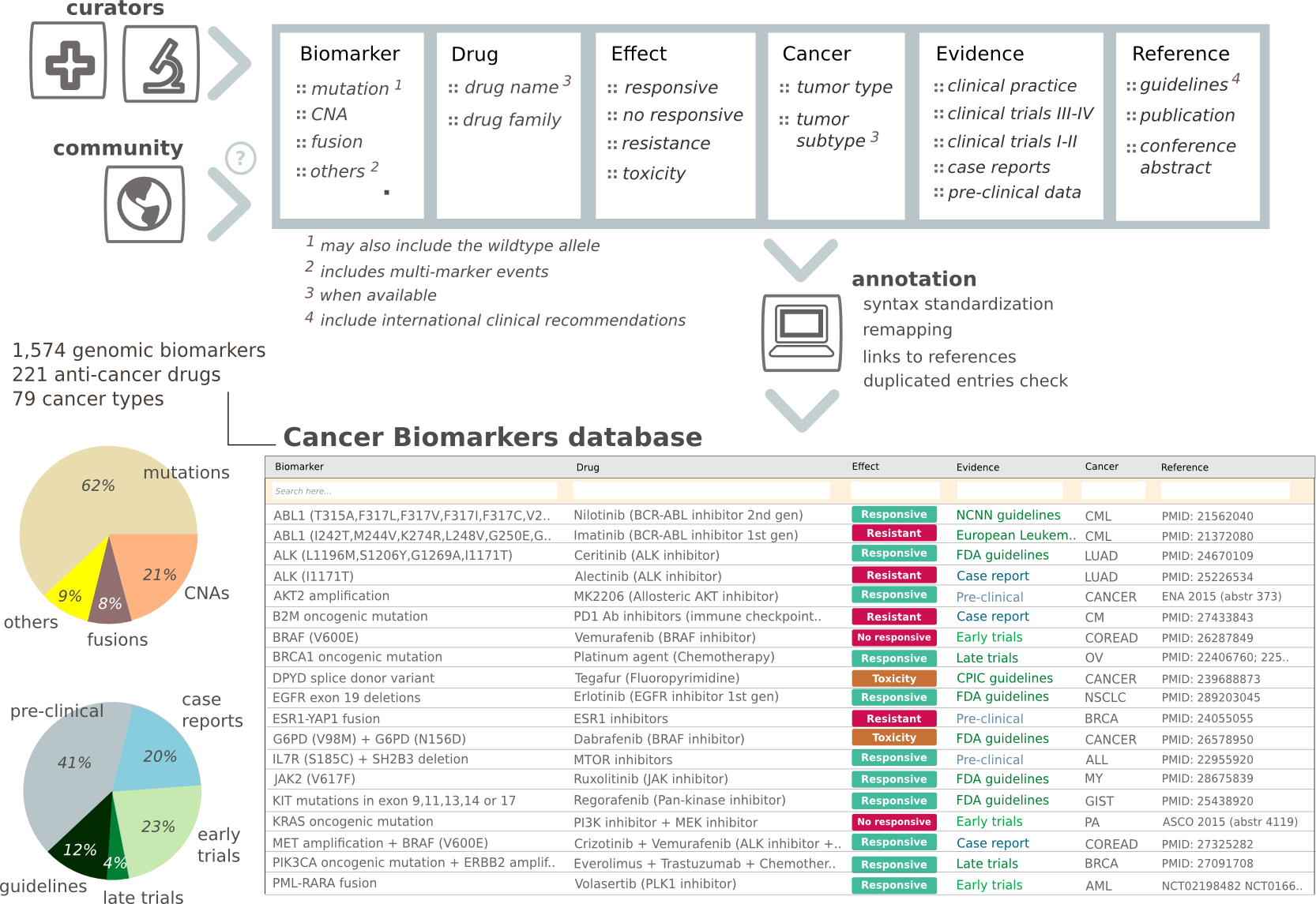
Schema of the Cancer Biomarkers database
What other resources are used to identify potentially actionable events?
Biomarkers of drug response integrated within the Variant Interpretation for Cancer Consortium Meta-Knowledgebase, collected from 2 stable and maintained resources (CIViC and OncoKB) are also used in the CGI to identify potentially actionable events in an individual's tumor. Please, note that we obtain CiVIC and OncoKB annotations from the VICC database, the contents of which may show some delay with respect to that of the original resources. In any case, links to the content of the original databases are provided for each biomarker.
Can CGI be used to interpret more than one tumor sample at a time?
A CGI execution can include the alterations detected in one or more tumor samples, as far as they correspond to the same cancer type. To distinguish the results of several samples you need to provide one column in the mutation file with a sample identifier.
Why is it necessary to select a cancer type when interpreting a sample(s)?
BoostDM models used to interpret the mutations are specific for a cancer gene in a tumor type, reflecting mechanisms of tumorigenesis that may vary between tissues. The selection of the tumor type of the sample under interpretation is thus key to choose the model specific for the tumor type in question, or the closest model if a specific one is not available. The interpretation of mutations via OncodriveMUT also employs knowledge on the role of each gene and gene region in specific cancer types, obtained from the analysis of large sequenced cohorts of tumors. Moreover, the identification of known driver alterations as well as the biomarkers of drug response take into account the match between the tumor type in which they have been described and the one of the sample under analysis.
Why is there a cancer taxonomy?
The cancer taxonomy (or oncotree) is important for the selection of general BoostDM models that are appropriate to interpret the mutations of a cancer gene in a tumor type for which a specific model is not available. For instance, if no specific model for a gene in lung squamous carcinoma --the tumor type selected by the user-- is available, then a model trained pooling the mutations observed across different histological types of non-small cell lung cancer may be used.
What is the format of the CGI output?
When the CGI is run via the web, its results are provided via interactive reports. These web reports can be interactively browsed and configured by the user. An alterations analysis report contains the annotations of all variants, which empowers the user's review, and the labels for those known or predicted to be drivers by BoostDM or OncodriveMUT. A biomarkers report contains the putative biomarkers of drug response found in the tumor organized according to distinct levels of clinical relevance. When run in the command line, these two reports are provided as tab-separated flat files.
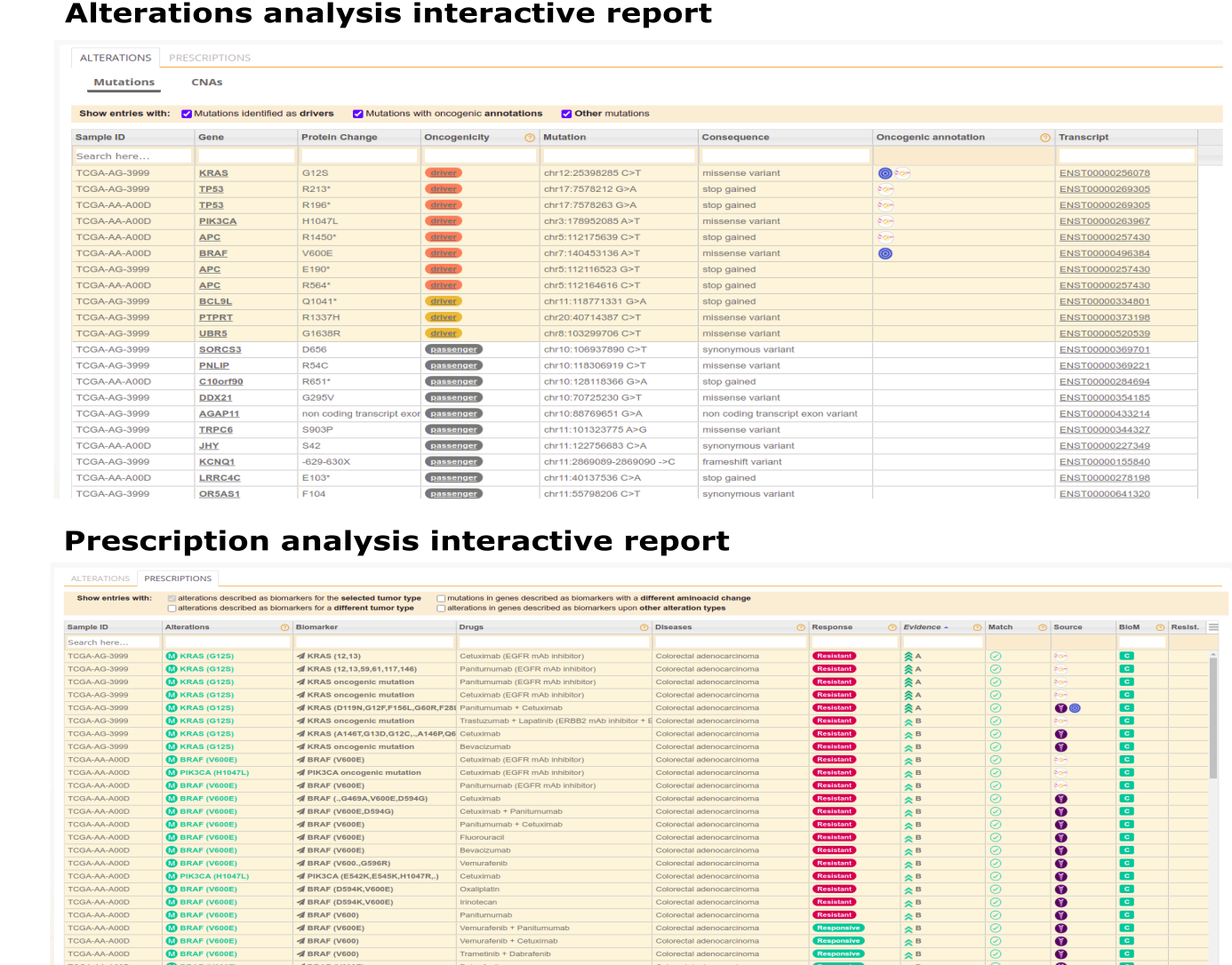
CGI output example
Which is the CGI license?
The results generated by the CGI output are under a Creative Commons Attribution-NonCommercial 4.0 (BY-NC) license.
How to cite CGI?
When referring to the datasets, any result generated by the CGI framework or the CGI resource itself, please cite: In silico saturation mutagenesis of cancer genes. doi: https://doi.org/10.1101/140475;
Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations; doi: https://doi.org/10.1101/140475
The main page https://www.cancergenomeinterpreter.org.
What is the input to run the CGI analysis?
The input (either pasted in the analysis box or uploaded within a file) is a list of alterations detected in one or more tumor samples. The analysis is always run on each sample separately, even if the user inputs mutations in more than one tumor. The alterations may include point mutations, copy number alterations and translocations (see format) with one alteration per line. If uploaded, the alterations may be separated across several files, which will be pooled before analysis.
Importantly, only mutations affecting the aminoacid sequence of protein-coding genes are analyzed by CGI. Thus, if you submit whole-genome mutations, the vast majority of them will be ignored by the system and generate warning messages by failure to map to exons.
Which mutations formats are accepted?
Check our format help page.
Which human genome assemblies are accepted?
Mutations mapped to either the GRCh37 or GRCh38 assemblies of the human genome are accepted. The user needs to select the assembly of the mutations in the analysis page.
How long does it take to have my CGI analysis finished?
The execution time depends on i) how long the job takes to get a slot in the cluster, ii) the time required by the data structures used by the CGI to be loaded, and iii) the number of entries to be analyzed. Therefore, to get the results may take a while even if only a few mutations are submitted. If you want to have an overview of the CGI results, you can check out the examples
Are there any platform requirements for the mutation data?
CGI interprets mutations obtained using any sequencing platform (e.g. exome sequencing, whole-genome sequencing or a gene panel) or calling pipeline. Of note, if the data include germline variants (i.e. the mutations have been not called only for somatic events), this can add some noise to the CGI driver analysis, although the oncogenic events (both known and predicted) are expected to be highly enriched for true somatic mutations. Also, germline variants that are known as cancer related (e.g. BRCA variants that predispose to breast tumors) are expected to be detected by the CGI. Of note, common polymorphisms (AF>1%) identifications are included in the CGI.
What happens if a mutation can not be mapped to the reference genome?
Mutations that cannot be mapped (if any) will be filtered out from the CGI analysis. Reasons of incorrect mapping include an unrecognizable format or incorrect mutation information (e.g. an invalid reference allele or chromosome position). If the number of unmapped mutations exceeds 30% of all mutations submitted, the system assumes that the data are not reliable and does not proceed with the interpretation. We also remove mutations mapping to non-exonic regions, with the exception of potential splice-site affecting mutations. Of note, unmapped mutations are accessible as a separate file for further review.
Are any mutations not analyzed/included in the report?
CGI interprets all exonic and/or potentially protein sequence-affecting mutations.
Do I need to login to run a CGI analysis?
The CGI can be used --including running analyses-- without login. The results of the run will be available for 24 hours at the link provided by the system upon submission. On the other hand, if the user logs into the system and clicks the button 'Save' in the analysis box, it won't be removed after 24h. Note that the login process only requires a valid email address and access is immediately granted. Any analyses launched with a logged user are automatically saved.
Can I download CGI results as a file?
Yes, once the interpretation is completed, the results can be downloaded as tab-separated files through the corresponding button.
Are my CGI results private?
To prevent unauthorized access or disclosure, to maintain data accuracy, and to ensure the appropriate use of information, CGI uses a range of reasonable physical, technical, and administrative measures to safeguard your Personal Information, in accordance with current technological and industry standards. In particular, all connections to and from our website are encrypted using Secure Socket Layer (SSL) technology. CGI never has access to your password and uses a trusted third party (gmail, mozilla persona, yahoo...) protocol to authenticate you. While the analyses are running and accessible from our web server they are stored in our private servers. When you remove an analysis, it will be completely and permanently removed from our servers, and we do not keep any copy. We do not share any files with third parties.
Can I share my CGI results?
Yes. Once the analysis is completed, the user may share the results by clicking the 'Share' button. But be aware that anyone with the URL will be able to access your analysis then.
Can I provide feedback on CGI?
For any comment, suggestion or error report, please contact with bbglab@irbbarcelona.org
Is there a REST API?
Yes, find all the details in this link: www.cancergenomeinterpreter.org/rest_api
How to access old versions of the CGI
CGI 2018 (the previous version of the tool) may be accessed here: https://www.cancergenomeinterpreter.org/2018